Main Second Level Navigation
Brenda Andrews
C.C., PhD, FRSC

Qualification
- University of California, San Francisco, CA, U.S., Research Fellow with Dr. Ira Herskowitz, 1991.
- University of Toronto, PhD in Medical Biophysics, 1986.
- University of Toronto, BSc in Zoology, 1980.
MY RESEARCH OVERVIEW (GO TO SCIENTIFIC OVERVIEW)
Building maps of cell function
It’s amazing to think that a single-celled organism could unlock the secrets to the complexity of human health and disease.
All disease has a genetic basis, whether in genes inherited by the affected individual, environmentally-induced genetic changes that produce a cancer, or the genes of a virus and their interaction with an infected person. But, despite astounding advances in biomedical research, the cause of common human diseases remains largely unknown. Common disorders have proven to be difficult to study as they likely arise from a complex interplay of many different genes. The importance of genetic interactions in disease has been emphasized by genome sequencing projects, particularly with the revelation of the human genome sequence, which estimates 4.1 million variants - differences in DNA - between any two people! Determining how combinations of genetic variants or perturbations manifest themselves, particularly in the context of human disease, is a formidable challenge. To address this challenge, we are collaborating with Charlie Boone’s group to understand how genes interact. To do this, we are using a simple model system, the single-celled budding yeast; yeast is not only famous to beer- and wine-makers, but is also celebrated as a premier model system for understanding the basics of human biology.
We are also studying how cells divide. It has become clear that the machinery that controls cell division is universally subverted in cancer cells. We aim to understand how the cell cycle is regulated, with the overarching goal of contributing to the world-wide effort to design better cancer therapies. Research in our lab is supported by the Canadian Institute of Health Research, National Institute of Health, the Ontario Research Fund and the Canadian Institute for Advanced Research (CIFAR).
The Donnelly Centre is comprised of leading genomics and proteomics researchers in a single location. The research atmosphere is very dynamic and open, fostering a creative and collaborative spirit. Building-wide, weekly seminars provide a platform to share our most recent work with fellow scientists from various fields including computer science and bioinformatics, chemistry, engineering and biology. These seminars are often the setting in which to craft new ideas and creative ways to solve difficult biological problems.
SCIENTIFIC RESEARCH OVERVIEW
We are interested in understanding cellular signaling processes and genetic networks in budding yeast using functional genomics, high content screening and gene expression tools.
Current projects in the lab include:
1. Functional Genomics and Genetic Networks
Our lab uses synthetic genetic array (SGA) technology (Tong et al., Science, 2001) combined with a variety of libraries of yeast strains to screen for genetic interactions. In collaboration with Dr. Charlie Boone’s group, we are investigating genetic redundancy by assessing how combinatorial mutations affect fitness, measured by colony size. We’ve created all pair-wise double mutant combinations for all 6000 yeast genes, creating the first complete genetic interaction map for a model eukaryotic cell. The map is rich in functional information, enabling precise prediction of gene function and providing insight into functional modules in the cell. We are now exploring deeper redundancy in biological networks by assessing triple mutant strains for genetic interactions, and by studying how genetic networks are rewired in response to environmental perturbations.
We also use SGA to probe interactions between enzymes and their targets to better understand signaling pathways that involve phosphorylation, acetylation and ubiquitination, for example.
2. Phenomics:
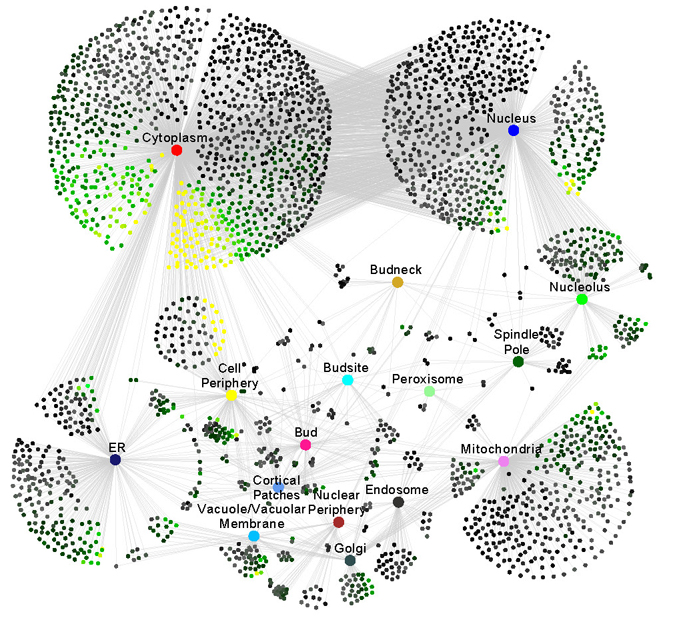
We have combined SGA with high content screening (HCS) to assess changes in protein localization and abundance under a variety of genetic, environmental and chemical stresses using rapid image acquisition and analysis. Using a collection of yeast strains with cytosolic RFP, each containing a different GFP-tagged protein, we have generated the first flux network (Chong et al., Cell, 2015) that charts how proteins change in abundance and localization in response to perturbation. We would next like to investigate how protein localization and abundance change at different stages of the cell cycle and when cells are dying.
In our Marker Project, the SGA-HCS pipeline is used to follow changes in subcellular morphology, such as changes to compartments involved in endocytosis or an increase in DNA damage foci. We would like to understand, at a subcellular level, any changes that result from deletion of non-essential genes. Further to this, we are also interested in any morphological changes that precede lethality in the context of genetic interactions or in temperature-sensitive mutants.
We are constantly working to improve computational analysis of our data by advancing neural network and machine deep-learning approaches, in collaboration with Brendan Frey’s group.
3. Cell cycle-regulated transcription
Our lab has pioneered the use of SGA for genome wide analysis of reporter gene-expression, particularly for studying transcriptional changes throughout the cell cycle. We have used the so-called Reporter-SGA (R-SGA) method to discover new mechanisms regulating histone gene expression during the cell cycle, and to systematically identify genetic perturbations that influence expression of a collection of reporter genes, covering all cell cycle stages.
In collaboration with Dr. David Botstein’s lab from Princeton University we are building an overexpression collection of yeast genes using titratable estradiol promoter, which enables a faster detection of dynamic changes in gene expression than the traditional GAL system.
SELECT PUBLICATIONS:
- Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Chong YT, Koh JL, Friesen H, Duffy K, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. Cell. 2015 Jun 4;161(6):1413-24.
- A cell cycle-regulated oscillator coordinates core histone gene transcription by histone acetylation. Christoph F. Kurat, Jean-Philippe Lambert, Julia Petschnigg, Helena Friesen, Tony Pawson, Adam Rosebrock, Anne-Claude Gingras, Jeffrey Fillingham, and Brenda J. Andrews. PNAS. 2014 Sep 30;111(39):14124-9.
- Exploring the yeast acetylome using functional genomics. Kaluarachchi Duffy S, Friesen H, Baryshnikova A, Lambert JP, Chong YT, Figeys D, Andrews B. Cell. 2012 May 11;149(4):936-48.
