What We Offer
We offer sequencing, library preparation, and other services. Take a look at what’s available at the DSC, and then proceed to Getting Started to take the first step in your project.
Sequencing
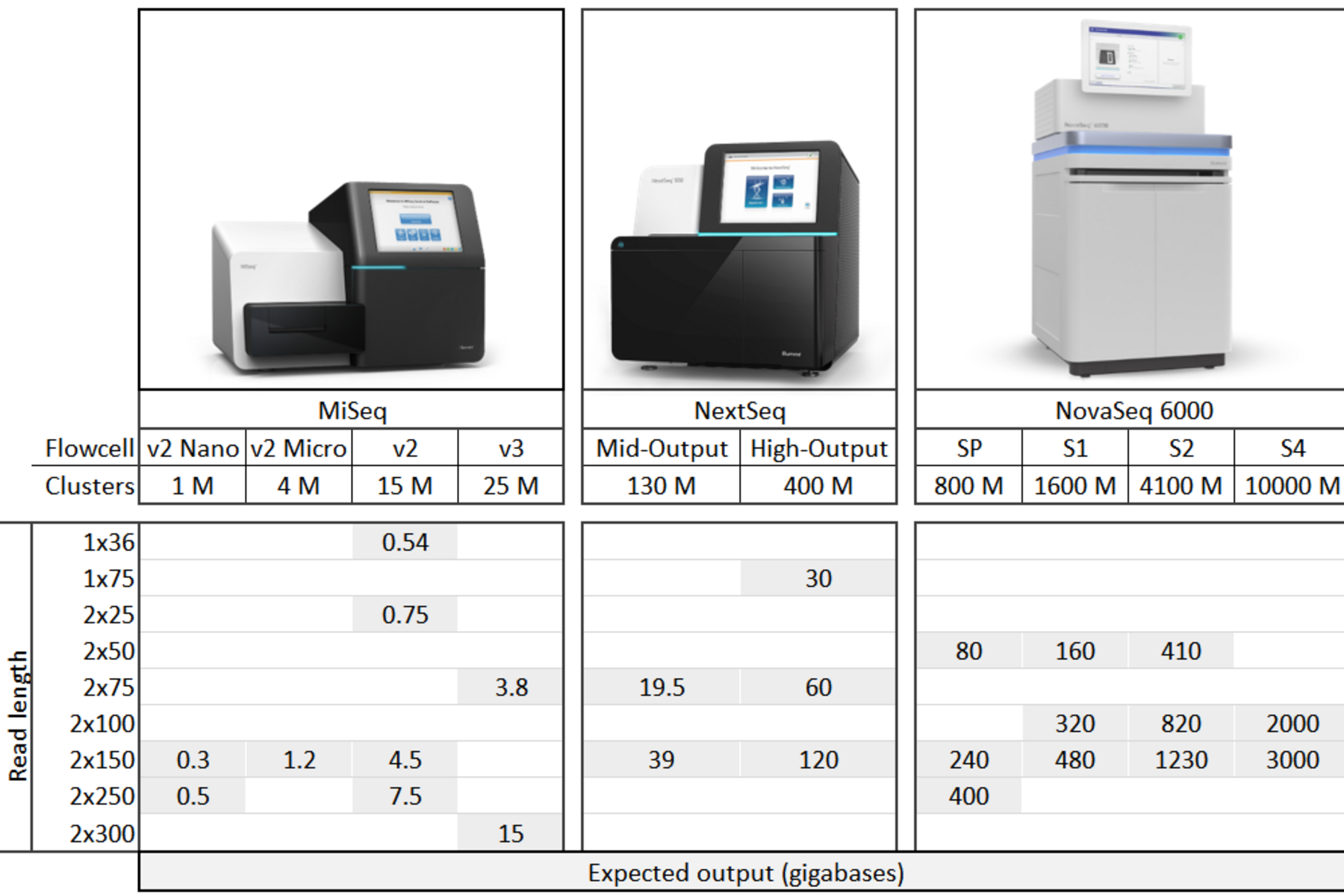
We accept libraries prepared by you and submitted directly to us for sequencing. Use the table below to explore which instrument best fits your project – grey cells indicate available run modes and Illumina’s expected data output in gigabases.
To help our users take advantage of the low cost/gigabase on the NovaSeq, we offer NovaSeq flex service, where we let you use part of a flowcell, and run your project alongside other compatible projects. Please note, however, that this can increase turnaround time as we wait for other projects – filling up a flowcell with your own samples is your best bet for getting data back quickly.
Library preparation
We also offer complete library preparation services for a wide variety of sample types and analysis needs.
DNA |
RNA |
|---|---|
|
DNA-Seq (e.g. shotgun) ChIP-Seq Methylation Amplicon sequencing Whole-exome sequencing |
Ribo-depletion RNA-Seq – stranded mRNA-Seq – stranded Small RNA-Seq mi RNA-Seq 3' end-Seq |
*Most kits listed here are kept in stock and available on-demand. For methylation sequencing, exome sequencing, and other methods not listed here, including Takara SMART chemistry, please contact us for a custom order. We're also happy to collaborate on method development for novel protocols.
Data distribution and storage
Sequence data are distributed on our servers as de-multiplexed compressed FASTQ files. Our default multiplex settings are trimming enabled, with 1 index mismatch allowed.
For users who are newer to NGS and bioinformatics, we recommend trying Illumina’s BaseSpace application. If you’ve signed up for an account, include your BaseSpace information on your Sample Information Sheet and we’ll upload your data directly there (timing dependent on network availability). This will allow you to make use of their many cloud-based sequencing analysis applications, including BWA, Cufflinks, TopHat, Velvet, Issac, RNAexpress, DNAstar, and FASTQC. Sign up and explore their toolset at Illumina.
Raw data (and samples) will be stored at the DSC for 3 months after project completion; demultiplexed fastq files are stored for two weeks.
Other services
Preparing libraries yourself but missing some key equipment? The DSC offers the following services:
-
DNA or RNA quantification: ThermoFisher Qubit (low-throughput) or Quant-iT (high-throughput), broad range or high sensitivity kits
-
RNA fragment analysis: Agilent TapeStation HS RNA ScreenTape or Bioanalyzer RNA Pico Chip
-
Library distribution analysis: Agilent TapeStation HS D1000 ScreenTape or BioAnalyzer HS DNA Chip
-
Library quantification: NEBNext Library Quant qPCR
-
Bead cleanups and size selection: MagBio HighPrep beads (Ampure XP equivalent)
Contact us about making use of other basic pieces of lab equipment (e.g. thermocycler, heating block, tube or plate centrifuge).
