Charles Boone
PhD, FRSC

Qualification
- University of Oregon, Oregon, OR, U.S., Research Fellow at Institute of Molecular Biology, 1989-1993.
- McGill University, PhD in Biology, 1989.
- Queen’s University, BSc in Chemistry and Math, 1982.
MY RESEARCH OVERVIEW (GO TO SCIENTIFIC OVERVIEW)
Global mapping of genetic and chemical-genetic interaction networks
Yeast, the microorganism underlying beer, bread, and wine, is providing spectacular insights into our general understanding of the biology of eukaryotic cells. Amazingly, the key components of its chromosomes and the function of many yeast genes are highly conserved in humans. While full genome sequences identify all genes within a particular organism, one of the next major challenges researchers are facing is to be able to explain the role of each gene and their functional organization within the cell. There are hundreds of yeast labs around the world and our collective goal is to develop a model a fundamental cell.
The yeast model system is famous for its relatively simple but elegant genetic methodology, often referred to as the “awesome power of yeast genetics.” Together with Dr. Brenda Andrews' lab, we developed an automated approach for yeast genetic analysis, called Synthetic Genetic Array (SGA) analysis, which enables us to make millions of different yeast strains carrying defined combinations of mutant genes. SGA maps gene-gene genetic interactions on an unprecedented scale, which allows us to determine which genes work together to control a particular biological processes, and we are testing all ~6000 yeast genes for genetic interactions within the context of 36 million double mutants.
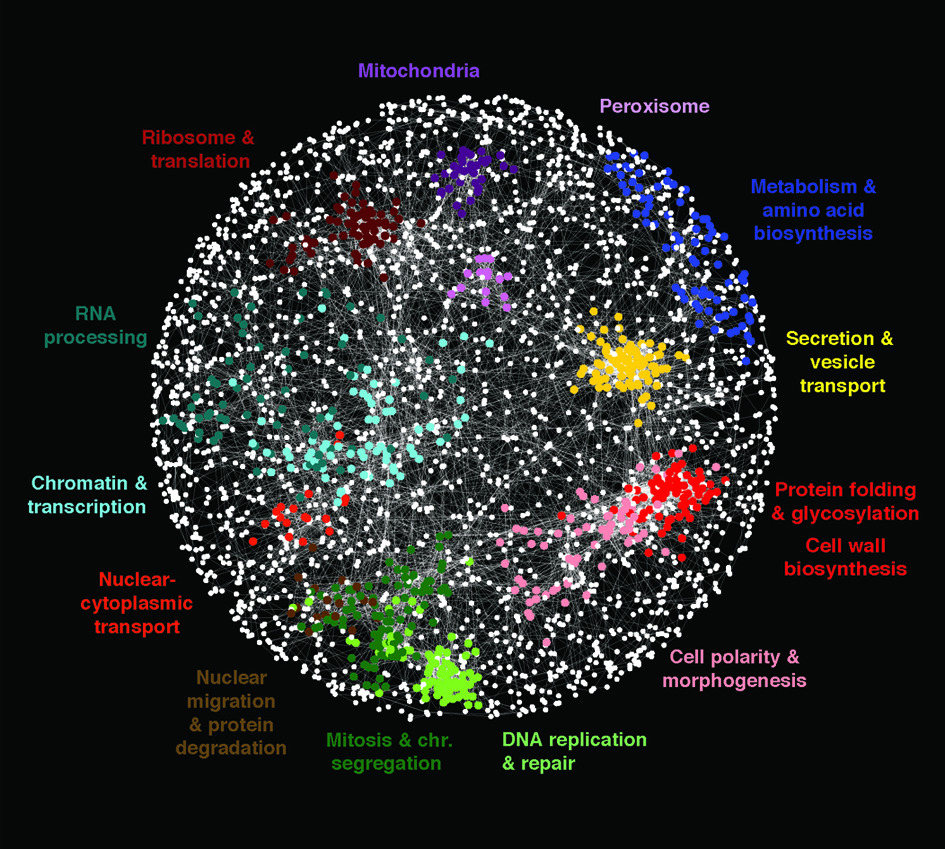
Our global yeast genetic network maps a functional wiring diagram of the cell. Importantly, what we observe about the control of cell growth and division in yeast is highly relevant to a molecular understanding of a number of diseases, including cancer. Moreover this global genetic network can be used to decipher the targets of bioactive molecules and lead drugs. Genetic networks may also provide insight into our understanding of the molecular basis of genetic diseases because gene-gene interactions appear to be powerful drivers of inherited phenotypes.
SCIENTIFIC RESEARCH OVERVIEW
In collaboration with Dr. Brenda Andrews' lab, we developed a technology called Synthetic Genetic Array (SGA) analysis, which allows us to combine multiple genetic elements in a single cell in a highly parallel manner. SGA takes advantage of using high-density yeast arrays composed of ~5000 viable deletion mutants or ~1000 temperatures sensitive (TS) alleles of essential genes, enabling us to carry out genome-wide genetic interactions screens. In particular, synthetic lethal genetic interactions, in which single mutants are viable but the corresponding double mutant is lethal, identifies genes that work together to control essential functions. To score these rare genetic interactions, we developed a robust method that relies on quantitative estimates of growth rate derived from colony size measurements, which allows for accurate measurement of both negative (e.g. synthetic lethal or sick) and positive (e.g. suppression) genetic interactions. Applying our high-throughput robotic pipeline has enabled the analysis of a complete set of yeast double mutants for genetic interactions quantitatively. Genetic networks derived from this analysis not only revealed detailed clusters of genes that represent specific pathways and complexes, but it provided a global view of the functional organization of a eukaryotic cell and facilitated discovery of roles for previously uncharacterized genes. Importantly, this work has defined the organizing principles of a global genetic interaction network, highlighting the incredible potential for genetic interactions to underlie inherited phenotypes and contribute to the genetic basis of diseases in humans.
Current projects in our lab include:
1. A Comprehensive Yeast Genetic Interaction Network
(i) SGA analysis with Essential Query Genes
A major goal of our research program is to complete the global yeast genetic interaction map by testing all ~18 million gene pairs for genetic interactions. In particular, we are now focused on the set of ~1,100 essential yeast genes, which are highly conserved and are enriched for core cellular machinery. We have constructed ~1000 conditional TS alleles of unique essential genes and tested them by SGA for genetic interactions. We found that essential genes are hubs on the global genetic network and incredibly rich in functional connections.
(ii) Duplicated Genes and Triple Mutant SGA Analysis
Derived from an ancient whole-genome duplication, there are ~400 pairs of nonessential duplicated genes. To quantify their functional redundancy and place these genes on the global network, we are conducting triple mutant SGA analysis using query strains carrying mutations in both paralog genes. Importantly, we have also extended this triple mutant analysis beyond duplicated genes in order to estimate the size and breadth of triple mutant genetic interaction space and begin to establish a general understanding the contribution of complex genetic interactions, involving more than two genes to phenotype heritability.
2. Large-scale Mapping of Mechanistically Diverse Genetic Interactions
To further address the genotype to phenotype problem, we are also generating new large-scale datasets that will reveal general systems-level rules associated with different types of genetic interactions that differ from those associated with two loss-of-function alleles. For example, we are mapping dosage suppression interactions for all yeast essential genes, in which the increase in copy number of one gene suppresses the fitness defect associated with a loss-of-function mutation in an essential gene. We are also conducting a large-scale analysis of genetic suppression, in which a mutation in one gene suppresses the phenotype of a mutation in another gene. Genetic suppression interactions are incredibly powerful because they are rich and functional information and also have direct therapeutic applications because a gene that can suppress the detrimental effects associated with a disease allele represents a putative drug target.
3. Chemical Genomics
We are developing a robust chemical-genomics platform designed to functionally annotate chemical libraries and link bioactive compounds to their cellular targets. Identifying compound-target relationships remains one of the rate-limiting steps in drug discovery. Scoring yeast mutants that are hypersensitive to a compound generates a diagnostic chemical-genetic profile indicative of a compound’s mode-of-action. Moreover, our global yeast genetic interaction network provides a key for interpreting chemical-genetic profiles and linking bioactive compounds to their targets. Our high-throughput approach allows us to screen large chemical libraries to functionally characterize thousands of compounds. Ultimately, these methods will define new tool compounds for probing biological systems and drug leads for human therapeutics.
4. Human Cancer Cell Genetic Interaction Maps
All of the general approaches we have developed for yeast are now applicable to human cells. Of course, similar global-level studies with human cells are more difficult and expensive but the complete yeast genetic interaction network should serve as a reference map to help develop strategies for mapping analogous networks in human cells in an informed manner. In collaboration with Dr. Jason Moffat's lab, we carried out one of the first large-scale genetic interaction maps in human cells and we are now building on this work using CRIPSR gene editing technology to map genetic networks in human cancer cells on a global scale. This work will be directly applicable to the Genotype to Phenotype problem in humans and synthetic lethal cancer therapies.
SELECT PUBLICATIONS
- Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Chong YT, Koh JL, Friesen H, Duffy K, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. Cell. 2015 Jun 4;161(6):1413-24.
- The genetic landscape of a cell. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, Prinz J, St.Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Li Z-Y, Liang W, Marback M, Paw J, San Luis B-J, Shuteriqi E, Tong AHY, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pál C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras A-C, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. Science. 2010. Jan 22; 327(5964):425-31.
- A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, Giaever G, Nislow C, Andrews B, Botstein D, Graham TR, Yoshida M, Boone C. Nat Biotechnol. 2009 Apr;27(4):369-77.
